fMRI volumes preprocessing
Table of contents
- Preprocessing pipeline
- 1 - Reference volume extraction
- 2 - Volumes co-registration
- 3 - Decoding preprocessing
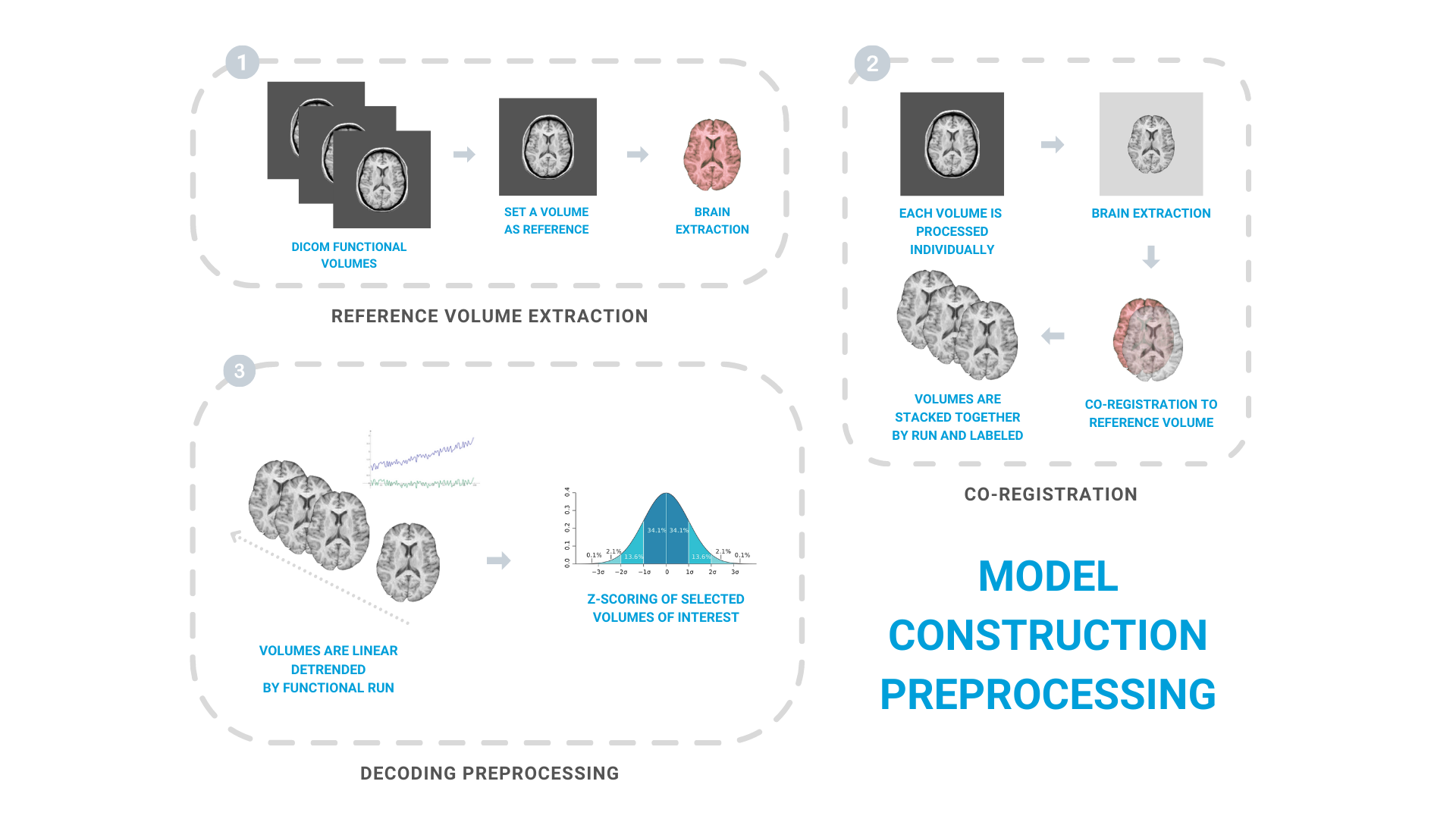
Preprocessing pipeline
Model construction session functional volumes are preprocessed using the same neuroimage software and preprocessing steps as they are preprocessed during real-time fMRI decoding neurofeedback training. This enables to maximize similarity between pipelines and avoid potential confusion variables.
As an starting point, a functional volume is set as reference volume to functional volumes of both model construction and neurofeedback training sessions.
Next, all functional volumes of the decoder construction session are individually corregistered to that reference volume and stacked together by functional run. All volumes are labeled using log files of experimental paradigm used during model construction session runs to identify each volume by its time from corresponding trial onset; its stimuli category; the experimental condition…
Once all functional volumes are labeled, volumes are linear detrended by fMRI run. Then, some volumes of interest are selected for model construction. Usually these are volumes falling within the Hemodynamic Response Function (H.R.F.) peak of each trial. Lastly, volumes of interest are Z-scored normalized at voxel level.
Preprocessing scripts of model construction session can be found in 1.model_construction/scripts/1.preprocessing sorted by required execution order.
Further, the pyDecNef pipeline provides example raw data (i.e., DICOM files) of a model construction session following recommended data structure already set in preprocessing scripts.
You just have to replace these data with yours, modify scripts to match your experimental settings, and you are ready to go.
1 - Reference volume extraction
NOTE:
Ensure that conda environment containing required dependencies for model construction
is activated, and dcm2niix and AFNI neuroimage software are preloaded
before using preprocessing scripts.
The first preprocessing pipeline step consists on selecting selecting a raw functional volume to serve as reference to all other volumes of model construction session, but also to volumes of neurofeedback training sessions.
Usually, fMRI runs start with the acquisition of a number of volumes (5 - 10 volumes) to allow for image stabilization that are then discarded.
In this pipeline, we select the first raw volume after MRI scanner heatup as reference image.
This DICOM volume is converted to NIfTI format (without compression) using dcm2niix to increase co-registration speed during real-time neurofeedback training sessions, and skull stripped using AFNI to facilitate co-registration to following volumes.
2 - Volumes co-registration
Raw volumes of all model construction runs are similarly converted NIfTI format, skull stripped and co-registered using rigid body (6 parameter) transformations to the reference volume by using AFNI 3dvolreg program.
Heptic polynomial interpolation (7th orders polynomial) is set as default spatial interpolation method in the co-registration procedure.
For each subject, one can increase co-registration accuracy between images by manipulating erode or clfrac parameters in brain extraction preprocessing step. This will erode the brain mask inwards avoiding skull and tissue fragments alter co-registration.
NOTE: if you decide to change the default erode and clfrac parameters for a participant, is important that new parameters are also set within the corregistration pipeline function used during real-time neurofeedback training for that subject (i.e., corregistration_pipeline.py). Otherwise, real-time co-registration might not be as accurate as during model construction session.
Once all raw volumes of model construction session were independently co-registered to the reference volume, we can stack them together by each run.
….. LABELING
3 - Decoding preprocessing
After labeling co-registered volumes, these are prepared for the machine learning model training.
Specifically, all functional volumes are linearly detrended by fMRI run at voxel-level. Then, volumes of interest of all runs (i.e., volumes falling within H.R.F. peak of trials) are extracted and stacked together.
Lastly, to improve model performance between fMRI sessions, BOLD activity is scaled at voxel level to have mean of 0 and a standard deviation of 1 by using Z-scoring normalization.